Hydrogen sulfide regulates muscle RING finger-1 protein S-sulfhydration at Cys44 to prevent cardiac structural damage in diabetic cardiomyopathy
- PMID: 30734268
- PMCID: PMC7024715
- DOI: 10.1111/bph.14601
Hydrogen sulfide regulates muscle RING finger-1 protein S-sulfhydration at Cys44 to prevent cardiac structural damage in diabetic cardiomyopathy
Abstract
Background and purpose: Hydrogen sulfide (H2 S) plays important roles as a gasotransmitter in pathologies. Increased expression of the E3 ubiquitin ligase, muscle RING finger-1 (MuRF1), may be involved in diabetic cardiomyopathy. Here we have investigated whether and how exogenous H2 S alleviates cardiac muscle degradation through modifications of MuRF1 S-sulfhydration in db/db mice.
Experimental approach: Neonatal rat cardiomyocytes were treated with high glucose (40 mM), oleate (100 μM), palmitate (400 μM), and NaHS (100 μM) for 72 hr. MuRF1 was silenced with siRNA technology and mutation at Cys44 . Endoplasmic reticulum stress markers, MuRF1 expression, and ubiquitination level were measured. db/db mice were injected with NaHS (39 μmol·kg-1 ) for 20 weeks. Echocardiography, cardiac ultrastructure, cystathionine-γ-lyase, cardiac structure proteins expression, and S-sulfhydration production were measured.
Key results: H2 S levels and cystathionine-γ-lyase protein expression in myocardium were decreased in db/db mice. Exogenous H2 S reversed endoplasmic reticulum stress, including impairment of the function of cardiomyocytes and structural damage in db/db mice. Exogenous H2 S could suppress the levels of myosin heavy chain 6 and myosin light chain 2 ubiquitination in cardiac tissues of db/db mice, and MuRF1 was modified by S-sulfhydration, following treatment with exogenous H2 S, to reduce the interaction between MuRF1 and myosin heavy chain 6 and myosin light chain 2.
Conclusions and implications: Our findings suggest that H2 S regulates MuRF1 S-sulfhydration at Cys44 to prevent myocardial degradation in the cardiac tissues of db/db mice.
Linked articles: This article is part of a themed section on Hydrogen Sulfide in Biology & Medicine. To view the other articles in this section visit http://onlinelibrary.wiley.com/doi/10.1111/bph.v177.4/issuetoc.
© 2019 The British Pharmacological Society.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

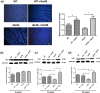



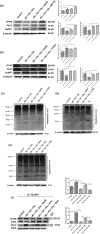
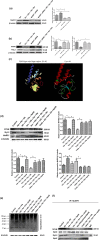
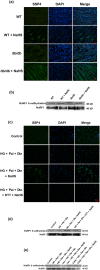
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

