Molecular pathway profiling of T lymphocyte signal transduction pathways; Th1 and Th2 genomic fingerprints are defined by TCR and CD28-mediated signaling
- PMID: 22413885
- PMCID: PMC3355027
- DOI: 10.1186/1471-2172-13-12
Molecular pathway profiling of T lymphocyte signal transduction pathways; Th1 and Th2 genomic fingerprints are defined by TCR and CD28-mediated signaling
Abstract
Background: T lymphocytes are orchestrators of adaptive immunity. Naïve T cells may differentiate into Th1, Th2, Th17 or iTreg phenotypes, depending on environmental co-stimulatory signals. To identify genes and pathways involved in differentiation of Jurkat T cells towards Th1 and Th2 subtypes we performed comprehensive transcriptome analyses of Jurkat T cells stimulated with various stimuli and pathway inhibitors. Results from these experiments were validated in a human experimental setting using whole blood and purified CD4+ Tcells.
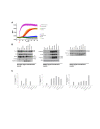
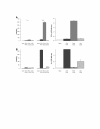
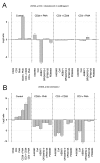
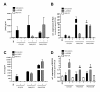
Results: Calcium-dependent activation of T cells using CD3/CD28 and PMA/CD3 stimulation induced a Th1 expression profile reflected by increased expression of T-bet, RUNX3, IL-2, and IFNγ, whereas calcium-independent activation via PMA/CD28 induced a Th2 expression profile which included GATA3, RXRA, CCL1 and Itk. Knock down with siRNA and gene expression profiling in the presence of selective kinase inhibitors showed that proximal kinases Lck and PKCθ are crucial signaling hubs during T helper cell activation, revealing a clear role for Lck in Th1 development and for PKCθ in both Th1 and Th2 development. Medial signaling via MAPkinases appeared to be less important in these pathways, since specific inhibitors of these kinases displayed a minor effect on gene expression. Translation towards a primary, whole blood setting and purified human CD4+ T cells revealed that PMA/CD3 stimulation induced a more pronounced Th1 specific, Lck and PKCθ dependent IFNγ production, whereas PMA/CD28 induced Th2 specific IL-5 and IL-13 production, independent of Lck activation. PMA/CD3-mediated skewing towards a Th1 phenotype was also reflected in mRNA expression of the master transcription factor Tbet, whereas PMA/CD28-mediated stimulation enhanced GATA3 mRNA expression in primary human CD4+ Tcells.
Conclusions: This study identifies stimulatory pathways and gene expression profiles for in vitro skewing of T helper cell activation. PMA/CD3 stimulation enhances a Th1-like response in an Lck and PKCθ dependent fashion, whereas PMA/CD28 stimulation results in a Th2-like phenotype independent of the proximal TCR-tyrosine kinase Lck. This approach offers a robust and fast translational in vitro system for skewed T helper cell responses in Jurkat T cells, primary human CD4+ Tcells and in a more complex matrix such as human whole blood.
Figures




References
-
- Brogdon JL, Leitenberg D, Bottomly K. The potency of TCR signaling differentially regulates NFATc/p activity and early IL-4 transcription in naive CD4+ T cells. J Immunol. 2002;168:3825–3832. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

