A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability
- PMID: 19647519
- PMCID: PMC2772893
- DOI: 10.1016/j.molcel.2009.06.021
A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability
Abstract
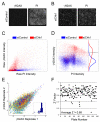
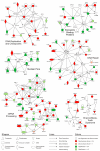
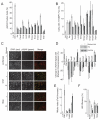
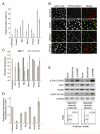
Signaling pathways that respond to DNA damage are essential for the maintenance of genome stability and are linked to many diseases, including cancer. Here, a genome-wide siRNA screen was employed to identify additional genes involved in genome stabilization by monitoring phosphorylation of the histone variant H2AX, an early mark of DNA damage. We identified hundreds of genes whose downregulation led to elevated levels of H2AX phosphorylation (gammaH2AX) and revealed links to cellular complexes and to genes with unclassified functions. We demonstrate a widespread role for mRNA-processing factors in preventing DNA damage, which in some cases is caused by aberrant RNA-DNA structures. Furthermore, we connect increased gammaH2AX levels to the neurological disorder Charcot-Marie-Tooth (CMT) syndrome, and we find a role for several CMT proteins in the DNA-damage response. These data indicate that preservation of genome stability is mediated by a larger network of biological processes than previously appreciated.
Figures


References
-
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. - PubMed
-
- Azzalin CM, Lingner J. The human RNA surveillance factor UPF1 is required for S phase progression and genome stability. Curr Biol. 2006;16:433–439. - PubMed
-
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. - PubMed
-
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

