Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae
- PMID: 17475768
- PMCID: PMC1924816
- DOI: 10.1091/mbc.e06-12-1149
Accumulation of polyadenylated mRNA, Pab1p, eIF4E, and eIF4G with P-bodies in Saccharomyces cerevisiae
Abstract
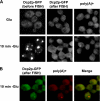
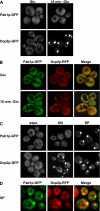
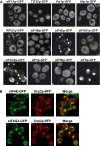
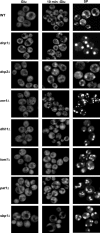
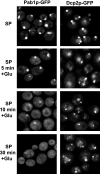
Recent experiments have shown that mRNAs can move between polysomes and P-bodies, which are aggregates of nontranslating mRNAs associated with translational repressors and the mRNA decapping machinery. The transitions between polysomes and P-bodies and how the poly(A) tail and the associated poly(A) binding protein 1 (Pab1p) may affect this process are unknown. Herein, we provide evidence that poly(A)(+) mRNAs can enter P-bodies in yeast. First, we show that both poly(A)(-) and poly(A)(+) mRNA become translationally repressed during glucose deprivation, where mRNAs accumulate in P-bodies. In addition, both poly(A)(+) transcripts and/or Pab1p can be detected in P-bodies during glucose deprivation and in stationary phase. Cells lacking Pab1p have enlarged P-bodies, suggesting that Pab1p plays a direct or indirect role in shifting the equilibrium of mRNAs away from P-bodies and into translation, perhaps by aiding in the assembly of a type of mRNP within P-bodies that is poised to reenter translation. Consistent with this latter possibility, we observed the translation initiation factors (eIF)4E and eIF4G in P-bodies at a low level during glucose deprivation and at high levels in stationary phase. Moreover, Pab1p exited P-bodies much faster than Dcp2p when stationary phase cells were given fresh nutrients. Together, these results suggest that polyadenylated mRNAs can enter P-bodies, and an mRNP complex including poly(A)(+) mRNA, Pab1p, eIF4E, and eIF4G2 may represent a transition state during the process of mRNAs exchanging between P-bodies and translation.
Figures




References
-
- Beelman C. A., Parker R. Differential effects of translational inhibition in cis and in trans on the decay of the unstable yeast MFA2 mRNA. J. Biol. Chem. 1994;269:9687–9692. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

