NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system
- PMID: 17244707
- PMCID: PMC1785264
- DOI: 10.1073/pnas.0609535104
NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system
Abstract
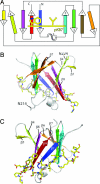
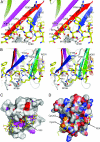
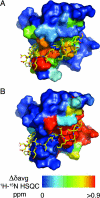
Type IV secretion (T4S) systems translocate DNA and protein effectors through the double membrane of Gram-negative bacteria. The paradigmatic T4S system in Agrobacterium tumefaciens is assembled from 11 VirB subunits and VirD4. Two subunits, VirB9 and VirB7, form an important stabilizing complex in the outer membrane. We describe here the NMR structure of a complex between the C-terminal domain of the VirB9 homolog TraO (TraO(CT)), bound to VirB7-like TraN from plasmid pKM101. TraO(CT) forms a beta-sandwich around which TraN winds. Structure-based mutations in VirB7 and VirB9 of A. tumefaciens show that the heterodimer interface is conserved. Opposite this interface, the TraO structure shows a protruding three-stranded beta-appendage, and here, we supply evidence that the corresponding region of VirB9 of A. tumefaciens inserts in the membrane and protrudes extracellularly. This complex structure elucidates the molecular basis for the interaction between two essential components of a T4S system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Schroder G, Lanka E. Plasmid. 2005;54:1–25. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

