Dual negative control of spx transcription initiation from the P3 promoter by repressors PerR and YodB in Bacillus subtilis
- PMID: 17158660
- PMCID: PMC1855716
- DOI: 10.1128/JB.01520-06
Dual negative control of spx transcription initiation from the P3 promoter by repressors PerR and YodB in Bacillus subtilis
Abstract
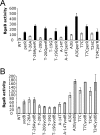
The spx gene encodes an RNA polymerase-binding protein that exerts negative and positive transcriptional control in response to oxidative stress in Bacillus subtilis. It resides in the yjbC-spx operon and is transcribed from at least five promoters located in the yjbC regulatory region or in the yjbC-spx intergenic region. Induction of spx transcription in response to treatment with the thiol-specific oxidant diamide is the result of transcription initiation at the P(3) promoter located upstream of the spx coding sequence. Previous studies conducted elsewhere and analyses of transcription factor mutants using transformation array technology have uncovered two transcriptional repressors, PerR and YodB, that target the cis-acting negative control elements of the P(3) promoter. Expression of an spx-bgaB fusion carrying the P(3) promoter is elevated in a yodB or perR mutant, and an additive increase in expression was observed in a yodB perR double mutant. Primer extension analysis of spx RNA shows the same additive increase in P(3) transcript levels in yodB perR mutant cells. Purified YodB and PerR repress spx transcription in vitro when wild-type spx P(3) promoter DNA was used as a template. Point mutations at positions within the P(3) promoter relieved YodB-dependent repression, while a point mutation at position +24 reduced PerR repression. DNase I footprinting analysis showed that YodB protects a region that includes the P(3) -10 and -35 regions, while PerR binds to a region downstream of the P(3) transcriptional start site. The binding of both repressors is impaired by the treatment of footprinting reactions with diamide or hydrogen peroxide. The study has uncovered a mechanism of dual negative control that relates to the oxidative stress response of gram-positive bacteria.
Figures



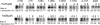


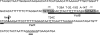
References
-
- Duwat, P., S. D. Ehrlich, and A. Gruss. 1999. Effects of metabolic flux on stress response pathways in Lactococcus lactis. Mol. Microbiol. 31:845-858. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

