One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas
- PMID: 16679460
- PMCID: PMC1475496
- DOI: 10.1105/tpc.105.038695
One of two alb3 proteins is essential for the assembly of the photosystems and for cell survival in Chlamydomonas
Abstract
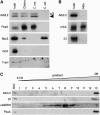
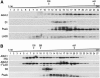
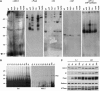
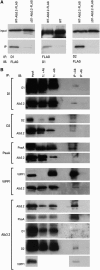
Proteins of the YidC/Oxa1p/ALB3 family play an important role in inserting proteins into membranes of mitochondria, bacteria, and chloroplasts. In Chlamydomonas reinhardtii, one member of this family, Albino3.1 (Alb3.1), was previously shown to be mainly involved in the assembly of the light-harvesting complex. Here, we show that a second member, Alb3.2, is located in the thylakoid membrane, where it is associated with large molecular weight complexes. Coimmunoprecipitation experiments indicate that Alb3.2 interacts with Alb3.1 and the reaction center polypeptides of photosystem I and II as well as with VIPP1, which is involved in thylakoid formation. Moreover, depletion of Alb3.2 by RNA interference to 25 to 40% of wild-type levels leads to a reduction in photosystems I and II, indicating that the level of Alb3.2 is limiting for the assembly and/or maintenance of these complexes in the thylakoid membrane. Although the levels of several photosynthetic proteins are reduced under these conditions, other proteins are overproduced, such as VIPP1 and the chloroplast chaperone pair Hsp70/Cdj2. These changes are accompanied by a large increase in vacuolar size and, after a prolonged period, by cell death. We conclude that Alb3.2 is required directly or indirectly, through its impact on thylakoid protein biogenesis, for cell survival.
Figures


References
-
- Barkan, A., and Goldschmidt-Clermont, M. (2000). Participation of nuclear genes in chloroplast gene expression. Biochimie 82 559–572. - PubMed
-
- Bartsevich, V.V., and Pakrasi, H.B. (1997). Molecular identification of a novel protein that regulates biogenesis of photosystem I, a membrane protein complex. J. Biol. Chem. 272 6382–6387. - PubMed
-
- Bonnefoy, N., Chalvet, F., Hamel, P., Slonimski, P.P., and Dujardin, G. (1994). OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J. Mol. Biol. 239 201–212. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

