Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice
- PMID: 11134350
- PMCID: PMC86640
- DOI: 10.1128/MCB.21.2.644-654.2001
Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice
Abstract
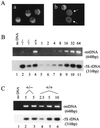
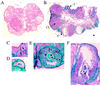
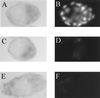
In vitro studies have implicated nuclear respiratory factor 1 (NRF-1) in the transcriptional expression of nuclear genes required for mitochondrial respiratory function, as well as for other fundamental cellular activities. We investigated here the in vivo function of NRF-1 in mammals by disrupting the gene in mice. A portion of the NRF-1 gene that encodes the nuclear localization signal and the DNA-binding and dimerization domains was replaced through homologous recombination by a beta-galactosidase-neomycin cassette. In the mutant allele, beta-galactosidase expression is under the control of the NRF-1 promoter. Embryos homozygous for NRF-1 disruption die between embryonic days 3.5 and 6.5. beta-Galactosidase staining was observed in growing oocytes and in 2. 5- and 3.5-day-old embryos, demonstrating that the NRF-1 gene is expressed during oogenesis and during early stages of embryogenesis. Moreover, the embryonic expression of NRF-1 did not result from maternal carryover. While most isolated wild-type and NRF-1(+/-) blastocysts can develop further in vitro, the NRF-1(-/-) blastocysts lack this ability despite their normal morphology. Interestingly, a fraction of the blastocysts from heterozygous matings had reduced staining intensity with rhodamine 123 and NRF-1(-/-) blastocysts had markedly reduced levels of mitochondrial DNA (mtDNA). The depletion of mtDNA did not coincide with nuclear DNA fragmentation, indicating that mtDNA loss was not associated with increased apoptosis. These results are consistent with a specific requirement for NRF-1 in the maintenance of mtDNA and respiratory chain function during early embryogenesis.
Figures



References
-
- Abbondanzo S J, Gadi I, Stewart C L. Derivation of embryonic stem cell lines. Methods Enzymol. 1993;225:803–855. - PubMed
-
- Barbacci E, Reber M, Ott M-O, Breillat C, Huetz F, Cereghini S. Variant hepatocyte nuclear factor 1 is required for visceral endoderm specification. Development. 1999;126:4795–4805. - PubMed
-
- Becker T S, Burgess S M, Amsterdam A H, Allende M L, Hopkins N. not really finished is crucial for development of the zebrafish outer retina and encodes a transcription factor highly homologous to human nuclear respiratory factor 1 and avian initiation binding repressor. Development. 1998;124:4369–4378. - PubMed
-
- Carmeliet P, Lampugnani M G, Moons L, Breviario F, Compernolle V, Bono F, Balconi G, Spagnuolo R, Oostuyse B, Dewerchin M, Zanetti A, Angellilo A, Mattot V, Nuyens D, Lutgens E, Clotman F, de Ruiter M C, Gittenberger-de Groot A, Poelmann R, Lupu F, Herbert J M, Collen D, Dejana E. Targeted deficiency or cytosolic truncation of the VE-cadherin gene in mice impairs VEGF-mediated endothelial survival and angiogenesis. Cell. 1999;98:147–157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
